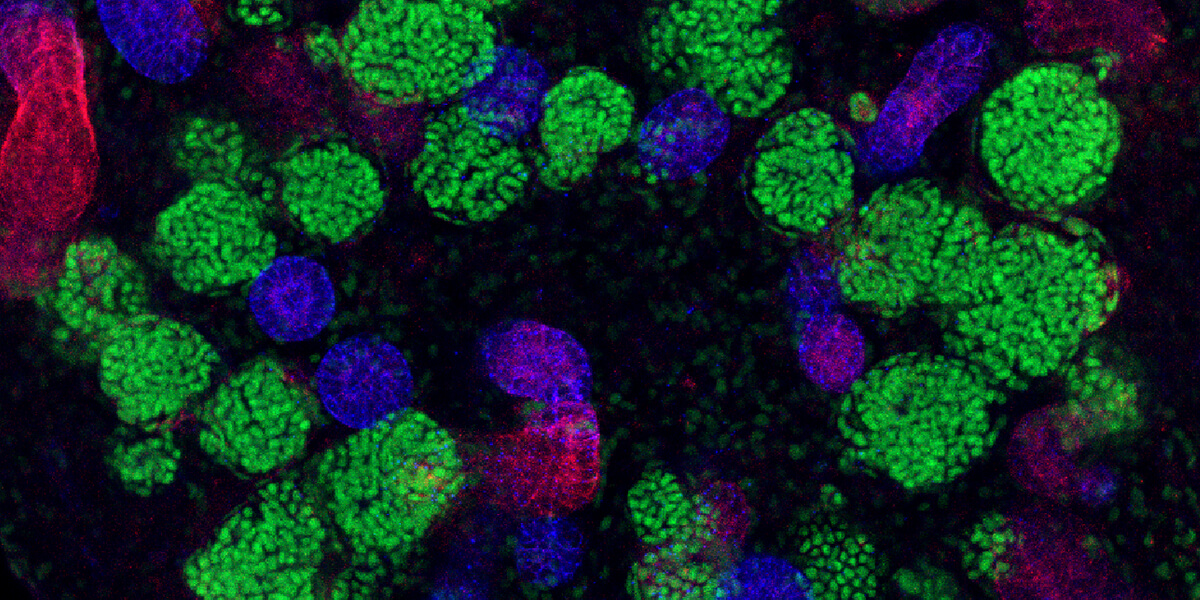
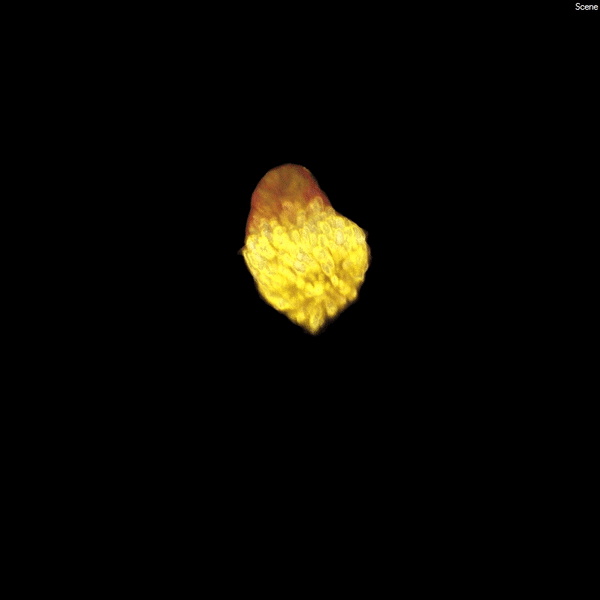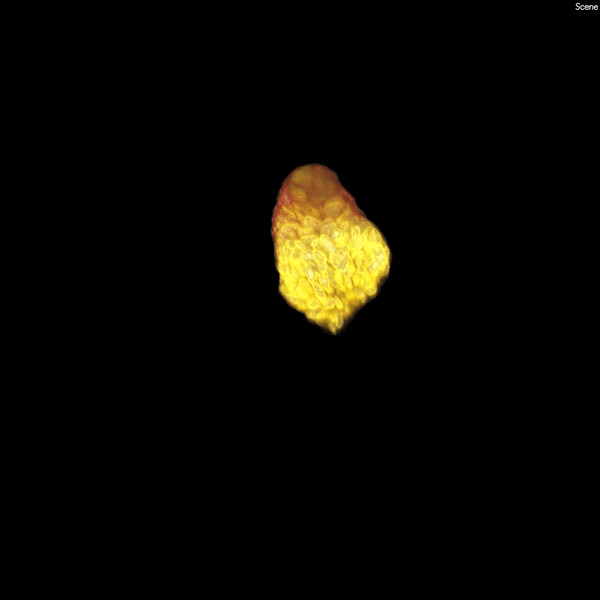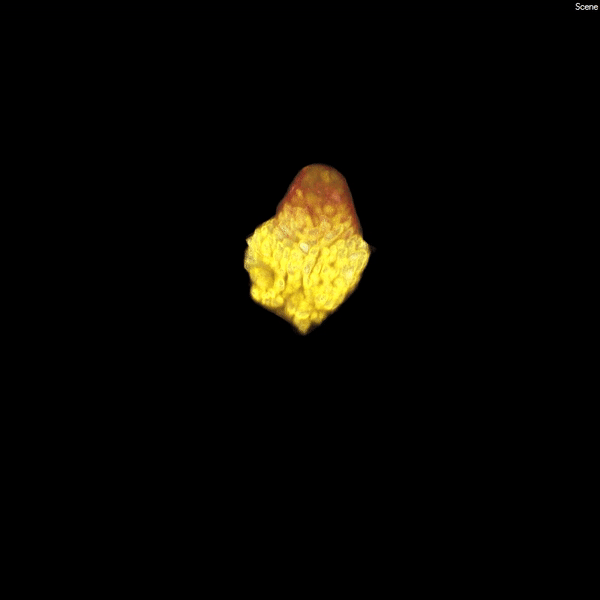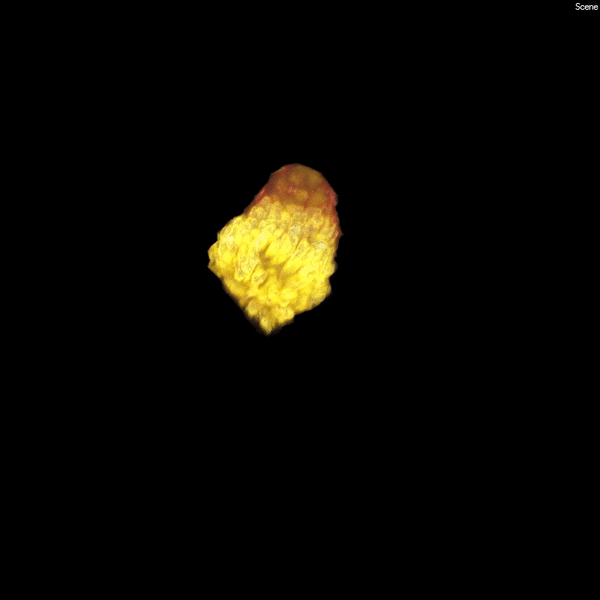
A kidney organoid on Day 16 of differentiation.A kidney organoid on Day 16 of differentiation. The staining depicts the segmentation of nephron-like structures in the organoid at an early developmental stage. Photo/Courtesy of Tracy Tran/Andy McMahon Lab, USC Stem Cell
A new USC study that reveals how kidneys form – which could pave the way for engineering new kidneys – was made possible in part by a collaboration with computer scientists at USC Viterbi’s Information Sciences Institute (ISI).
Over the past four years, USC Stem Cell researchers and computer scientists at ISI have been chronicling the molecular, cellular and genetic similarities and differences in how the complex kidney organ develops in mice and humans.
The team leveraged an innovative new software program, or “ecosystem of tools,” called DERIVA (Discovery Environment for Relational Information and Versioned Assets), developed by ISI division director Carl Kesselman and his team, to collect data from microscopes and organize them and other study data into virtual albums that can be easily shared with other researchers.
Each human kidney contains one million mature nephrons, which form an expansive tubular network (white) that filters the blood, ensuring a constant environment for all of our body’s functions. (Courtesy of Stacy Moroz and Tracy Tran/Andy McMahon Lab, USC Stem Cell)
“It’s a new way of doing science,” says Kesselman, a principal investigator at the USC Michelson Center for Convergent Bioscience, Dean’s professor in the Epstein Department of Industrial and Systems Engineering and professor of computer science in the Viterbi School of Engineering.
The study, part of a three-study series published on Feb. 15 in the Journal of the American Society of Nephrology, signifies a significant milestone in the development of treatment for kidney disease.
Understanding kidney development – specifically the function of nephrons, or filtering units – can allow medical researchers to understand, model and treat kidney disease, the 9th leading cause of death in the United States.
It is also an important step in making functioning kidneys for transplant, grown from a patient’s own tissue, a more realistic possibility in the future.
“Stem-cell based technologies hold great promise for developing kidney replacement and regeneration therapies,” said Nils Lindstrom, first author of the three new studies and a research associate in Stem Cell Biology and Regenerative Medicine at the Keck School of Medicine of USC.
“Getting there requires detailed knowledge of how kidneys normally form so the process can be replicated in cell cultures in the lab. Our data will help us and other scientists improve current techniques to make better tiny functional kidneys.”
A new way of doing science
Kidney disease is a complex problem, requiring a multidisciplinary approach. That’s why Andrew McMahon, study senior author and W.M. Keck Provost Professor of Stem Cell Biology and Regenerative Medicine and Biological Sciences at the Keck School of Medicine, teamed up with Kesselman, a study co-author.
This could be a gamechanger for the scientific research community.Andrew McMahon
By automating many of the tasks researchers needed to perform, such as recording data observed by a high-resolution microscope, the platform developed at ISI helped the scientists manage big scientific data, including experimental results, tissue samples, and gene expression data.
“The USC-created system allowed us to spend less time searching for elements and makes data easily accessible to the kidney community,” says McMahon. “This could be a gamechanger for the scientific research community.”
Kesselman likens DERIVA to the photo library on a cell phone— the software program gathers and organizes high-resolution images and other research data.
“At some point, you have so many photos that they become unmanageable. You can’t find the one you want. DERIVA automatically catalogs your data so you can analyze massive amounts of data with Zen-like calm and share them with all of your friends throughout the world.”

Professor Carl Kesselman. Photo/Damon Casarez.
The tool not only fast-tracked one of the three studies but also created an online, searchable library to aid other stem cell scientists in their kidney disease research.
The results will soon be made available to the global kidney research community as part of the National Institutes of Health-funded GenitoUrinary Development Molecular Anatomy Project (GUDMAP). Kesselman is also the principal investigator of GUDMAP, and ISI provides access to this data to researchers and clinicians across the globe.
Reproducible results
The research findings combined with the data resources provided by Kesselman and his team will help make human kidney development more accessible to the biomedical community.
“ISI researchers have been involved in every part of the process, from getting the data in the lab to allowing anyone interested in kidney development to access and reuse this data,” says Kesselman.
By consolidating all the documentation relating to a scientific study, from raw data to final results, the tool may also help to tackle the science community’s reproducibility crisis.
“We spends billions of dollars on research and we are discovering that many of the results are neither reproducible or reusable,” says Kesselman.
In fact, research suggests more than two thirds of scientists have tried and failed to reproduce another scientist’s experiments. One of the reasons may be that consolidating data, a must for ensuring reproducibility, takes an inordinate amount of time and effort.
Using DERIVA, instead of searching individually for each figure and data element, scientists can view, download, cite and share the data referenced in its full quality.
“In one click, readers have access to everything they need to explore the results and reproduce the experiments themselves,” says Kesselman.
“If any other researcher wants to use this data, they stand a much better chance. They know exactly what is available, and can interpret it right from the source.”
The research was supported by the National Institutes of Health (NIH DK107350, DK094526, DK110792, 5F32DK109616-02), National Institute of Diabetes and Digestive and Kidney Diseases (F31DK107216, 1U24DK110814), California Institute for Regenerative Medicine (CIRM LA1-06536) and Joint Medical/Research Council/Wellcome Trust (099175/Z/12/Z) as well as by a USC Stem Cell Hearst Fellowship and a USC Research Enhancement Fellowship.
Published on February 16th, 2018
Last updated on April 7th, 2025