Jenny Treweek, assistant professor of biomedical engineering, examines the physiological function of stress-related neuropeptide circuits using advanced neuroscience. Photo/Daniel Druhora
“There’s a brain in here!” Jenny Treweek exuberantly exclaims, raising a small, translucent tube containing the brain of a mouse that looks like it’s floating in Jell-O.
It’s her latest experiment in tissue clearing – one of her efforts to combine her background in chemistry, economics, systems neuroscience, optogenetics, biomedical engineering and even music to examine the physiological function of stress-related neuropeptide circuits involved in drug addiction, depression and PTSD.
Her research interests are summarized in this quote from famed molecular biologist Francis Crick that she has posted on her own webpage: “There is no scientific study more vital to man than the study of his own brain. Our entire view of the universe depends on it.”
The Gabilan Assistant Professor of Biomedical Engineering joined USC Viterbi in January 2019. We sat down with Treweek, an avid animal lover who spends time volunteering at animal shelters, to become more acquainted with her and her multifaceted research.
Tell us about the cat in the picture on your desk.
Oh, that’s Gracie. I could talk about her forever. I was a foster failure. This is ironic.
Actually, when I was applying for jobs, to recruit me, they showed me the (USC) Michelson Center for Convergent Bioscience, and it just blew me away. Finally, I thought, someone was doing science correctly, putting the electrical engineers with chemical engineers, with systems neuroscientists – people more like me – and that was a big draw. So, when they were talking to me about Michelson, I started researching – what is the Michelson building? Who is Michelson? You know, so I don’t sound like an idiot during interviews. And I look up a web page about Gary Michelson and it says that his other philanthropic interests, aside from biomedical sciences and research, include animal rescue. And so, I snapped a picture of that website and sent it to my sister asking her: “Courtney, who again is your benefactor at Found Animals Foundation?” And she said: “Oh, it’s Dr. Michelson.” So, now, he basically has both of my dad’s daughters gainfully employed.
And Gracie, my kitty, came from that cluster of animals at the Michelson Found Animals Foundation. My job was to get her up to two pounds and get her ready for adoption. But she had a severe health issue, something we coined “wilting kitty syndrome,” where they just don’t eat, they don’t gain weight. Her prognosis was not good. She would need chronic care for the rest of her life. At one point, her medical bills were costing me about $1,000 per month. It’s hard to saddle future adopters with this idea, so I decided that I would take care of her and so she came on board with me at the Michelson Center and now she’s going to turn six in November.
You have an interesting backstory. How did you become a scientist?
I was originally a chemist. I did my undergrad at Caltech in chemistry, economics and French. I had two wonderful advisors at Caltech: Rich Roberts, who is here now, and Peter Dervan. My first formative research experience was in Roberts’ lab where I was doing research alongside a grad student for about a year and a half at the end of my undergrad. Originally, I had planned to go into investment banking. It was the early 2000s and Caltech students were being poached by all the big banks. I had just signed on with Citigroup to become a financial analyst and I remember Peter Dervan calling me into his office and saying: “I heard you’re going to Citi Group, but if you were to have a backup plan, how would you think about grad school?” And he had sort of just laid out the path for me, I mean, to the point of looking up when I would take the GREs and had all my letters already ready to apply. He really went out on a limb, above the above and beyond the call of duty for just an advisor. And long story short, I ended up in chemistry grad school, doing my Ph.D. in chemistry at the Scripps Research Institute.
“I wondered, ‘How could smart, fully functioning people fall into the trap of becoming addicted to drugs? What is it that takes over?’”
Jennifer Treweek
But your research indicates that you’re not a classical chemist.
I’ve always been interested in neuroscience and had very biomedically driven projects. Lots of looking at small molecules, designing therapeutic antibodies that would bind small molecules that you could use in active or passive vaccination programs. And then you’re going from the design of these therapeutics into their preclinical testing and animal models. I already knew that I really liked the preclinical testing part, trying to understand why the molecules worked or why they didn’t. What about the biology was not being affected by the molecules you were delivering? And so, I decided to go into more neuroscience and biology driven research during my postdoc. This brought me back to Caltech to do a postdoc with Viviana Gradinaru within the biology and bioengineering department. She did absolutely beautiful work in showing how optogenetics, a technique that involves using light to control cells in living tissue, typically neurons that have been genetically modified to express light-sensitive ion channels, can be used to both discover pathways involved in Parkinson’s symptoms and then modulate those pathways to try and attenuate these symptoms.

In her lab, professor Jennifer Treweek, looks into the chemistry for tissue engineering and devising better ways to image tissue to obtain the most molecular information possible on that tissue. Photo/Daniel Druhora
Had you worked in optogenetics before?
No, but Viviana was very lovely, not just in opening my eyes to a new field; she taught me how to lead a conversation in a room as a young female scientist. I came to Viviana’s lab having done lots of animal testing at Scripps with Dr. George Koob, who is now the director of The National Institute on Alcohol Abuse and Alcoholism. His expertise was in alcohol and drug abuse, using animal models to characterize what leads to a maladaptive state of addiction: what are the brain circuits involved and how do you reverse it? So, I had absolutely no experience in optogenetics, but one of my long-term research interests was a neuropeptide that was very difficult to study with traditional methods and that’s CRF, or corticotropin-releasing factor. In a nutshell, it’s a peptide that triggers both the brain and the body’s response to stress.
What particularly attracted you to the study of stress related illnesses?
There’s lots of backstory as to why I was interested. Part of it would be that my dad fought in Vietnam, so, PTSD is obviously something I’m aware of. Also, working at Scripps in San Diego, there is a huge military presence and lots of my friends had come back from Afghanistan and were dealing with PTSD. The CRF system is obviously very important in stress related disorders like PTSD, generalized anxiety disorder and major depressive disorder. The other side of it was that during my undergrad I was very interested in music, and I had spent some time DJ’ing while in grad school. And I just remember being freaked out seeing people addicted to drugs in that scene and overdosing on drugs. I wondered, “How could smart, fully functioning people fall into the trap of becoming addicted to drugs? What is it that takes over?” Actually, while I was an undergrad, one the of very smartest M.D.- Ph.D. students I knew died from a methamphetamine overdose. It turns out the CRF system is intimately involved in this spiraling distress and that piqued my interest, but this neuropeptide system is very, very hard to study.
Why is it so hard to study the CRF system?
Much of my doctoral research was on therapeutics for cocaine overdose. To liken it, for example, to “Pulp Fiction,” where Uma Thurman ODs on heroin that she snorts — thinking it’s cocaine — and they start to administer a shot of adrenaline to the heart. That’s what you would be doing with cocaine — you would be administering a bolus shot of antibodies that would run through the body, bind up all the cocaine and pull that person out of an overdose. During this time, I was also looking at how these vaccination strategies could be useful for treating chronic states – to alleviate symptoms of withdrawal or to prevent relapse. Here, CRF is intimately involved in conveying the feelings of anxiety and dysphoria that well-up during drug withdrawal – symptoms which cause the addict to relapse. So obviously, compounds that target the CRF system, that modulate CRF signaling, could be invaluable therapeutically – either in combatting drug addiction, or in treating other stress-related disorders such as PTSD. But CRF, as a peptide, is not fun to work with when it comes to the medicinal chemistry side. Clinically useful peptide drugs are nontrivial to design in terms of their bioavailability and metabolic stability – plus they presumably need to get up to the brain. Alternatively, the synthetic steps that you would have to take to create a library of small-molecule CRF receptor antagonists would take a couple of years of work from a good organic chemist.
Then, let’s say you test your drug candidates in mice, and they absolutely don’t work. They don’t get into the brain, they have off target effects, they’re toxic. So, you could waste years and years of work just trying to make this panel of antagonists and be back at square one, and I didn’t want to spend my postdoc at a fume hood making compounds that potentially don’t work. Whereas with optogenetics you can specify the exact circuits in the brain (and in the body) that you want to turn on and off, and get real results about which circuits are relevant to disease symptoms, and how they should be modulated to confer therapeutic results. But, you also want to know the identity of the cells that comprise these circuits, and where these cells project throughout the brain… basically “who these cells are talking to and how loudly”. You want to be able to subdivide these circuits or these populations of cells based on their least common denominator, which might be a particular receptor or a particular transcript (i.e., genetic profile).. That’s when I got involved in tissue clearing and clarity.
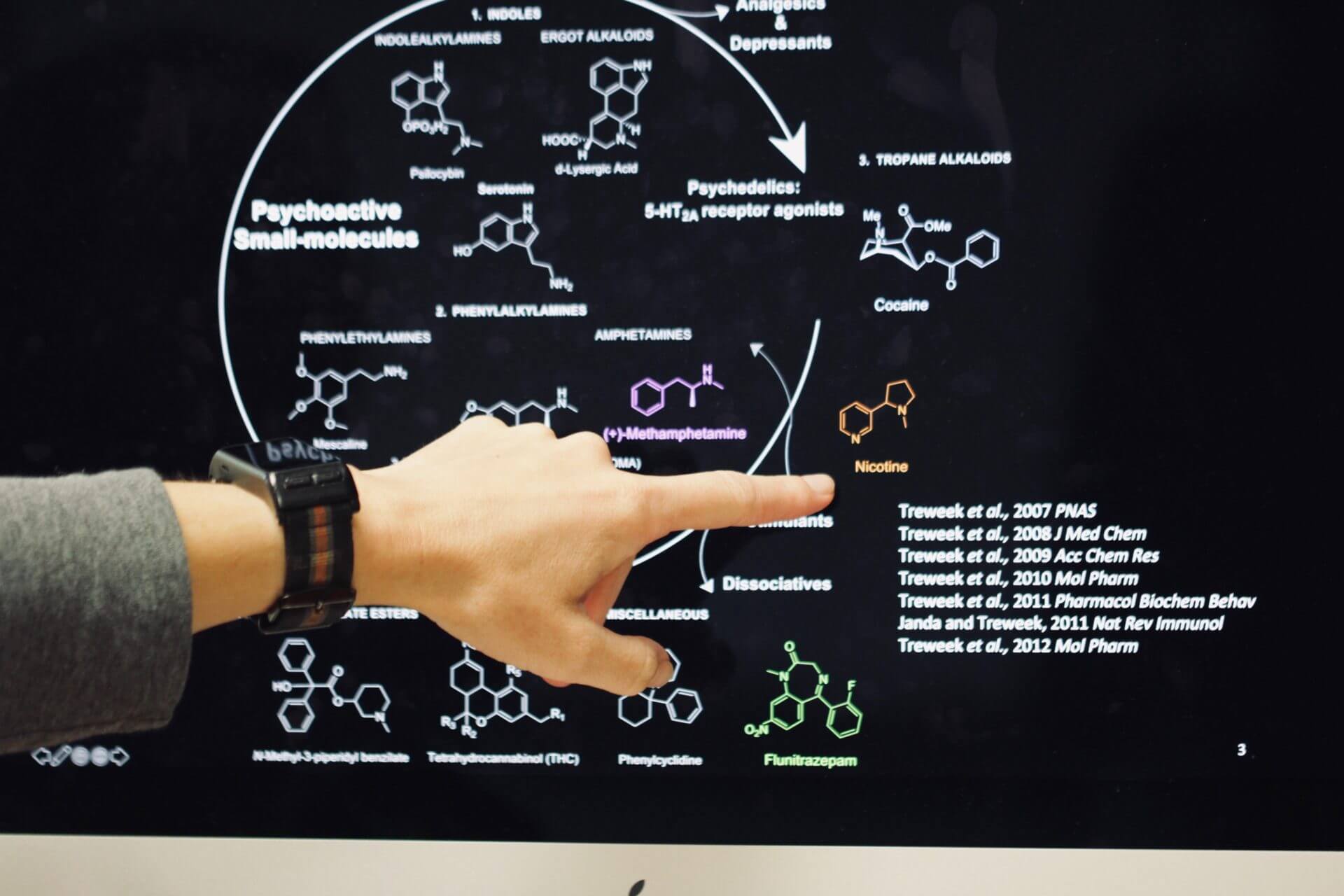
Jennifer Treweek explains the complexity of studying psychoactive small molecules with strong addictive effects such as nicotine. Photo / Daniel Druhora
Can you please describe what you mean by tissue clearing and clarity?
Okay, so in Viviana’s lab, we wanted to look at what cells were doing at an organ-wide scale. During Viviana’s doctoral work on Parkinson’s disease, it was important for her to record what cells were dying off on a brainwide scale. Viviana and her advisor, Karl Deisseroth at Stanford University, came up with the idea (termed “TEMPEST”) of filling up cells with a genetically-encoded polymer when they started to die so that after the cell was gone, the polymer scaffold would be like a permanent record of its existence – sort of like a police tape outline of a murder victim in the movies. In the case of Parkinson’s disease, where you have gross loss (80 percent is often cited) of midbrain dopamine neurons, a polymer record would allow you to reconstruct ex vivo where cells had once been – and perhaps provide a clue as to not only their former function, but as to why they were selectively lost during disease.
“I am much more interested not just in pharmacological strategies, but in identifying maladaptive circuit behaviors, and then envisioning new technologies that could modify the signaling patterns that give rise to disease symptoms.”
Jennifer Treweek
Taking this idea a step further, Kwanghun Chung, now a Professor at MIT, but formerly a chemical engineer in Karl Deisseroth’s lab, began to look at the utility of not just filling dying cells with polymer, but filling up all cells and also the extracellular spaces with a polymer. And, rather than just fill cells with an exogenous polymer that doesn’t really interact with the cell in any way, what if the polymer formed a very functional scaffold for the proteins within the tissue, so that we could stabilize the tissue and then perform chemistry on the tissue without damaging the biomolecular content? For the CLARITY technique, the idea is to embed the tissue in special type of polymer that’s a hydrogel – just like if you were to make Jell-O – that would be your hydrogel. And we started asking what if you could have a hydrogel that you perfuse throughout the brain that bound with all the proteins that are throughout the cells, then you could knock out the light scattering components, such as the fat or the lipids? What if you could remove all the layers that blocked you from seeing deep into the tissue and looking at its cellular content? So that’s CLARITY. There were some issues with CLARITY. One, it was very expensive, hard to do, and it risked damaging the tissue. And so, one of my projects, given my chemistry background, was to improve upon this initial CLARITY procedure to make it more user friendly and allow it to be paired with other applications.
Did that lead into what you are now doing here?
Yes, so one of the applications was labeling RNA transcripts and looking at not just protein because the original CLARITY was primarily to look at proteins and genetically-encoded fluorescent labels in tissue. What if we could also look at RNA transcripts within that clear tissue at the millimeter scale? We could look at the transcripts within a cell, but we would have the entire cell body there, as well as all of its cell neighbors, and we can see what far-off cells it’s “talking to” by also following its projections within that same piece of tissue. Now we have circuit information to go along with the cell’s protein and RNA content. And this is within the framework of an entire experimental pipeline – modulating cells that are alive with optogenetics or other techniques to observe their physiological relevance. Once I’m done with the in vivo experiments to, for example, model the symptoms of a stress-related disorder, I can take the tissues, label transcripts, clear the tissue and image changes, transcriptional changes that may have stemmed from the optogenetic modulation or disease pathology. In my case, this could involve identifying cells where CRF receptor up regulation or down regulation is prominent, and then quantifying the other transcriptional changes within those cells and looking at the downstream circuit effects. What does that mean within those circuits? What other cells are they talking to and how are they now “talking” differently? Although, we’ve done the initial validation of this experimental pipeline, we still have barriers to using it effectively for systems-neuroscience projects like coming up with better treatment strategies for drug addiction and depression.
And what specifically is your lab doing to address this challenge?
This is the experimental side of my lab. What would be a superior hydrogel formulation that would better stabilize the nucleic acid content of tissue that we could then promulgate in the brain to figure out the pathways for addiction? On the computational side, it’s basically a computational nightmare because we have point labels for single transcripts and now, we have to count these points in large sections of tissue. The algorithm is very feasible if you have a mono layer of tissue – you just have one image – and you just need your algorithm to decide on cell boundaries in a 2D plane, and then restrict the transcript counts to with each cell’s 2D boundary. But now, if we have a 3-D image stack, we have to be able to scan through each image of that large stack and do segmentation across 3-D cell boundaries across several of these images and then count the points within. So that’s much harder computationally and we’re working to address this problem in my lab.
What exactly are you hoping to solve with this?
In my case, I’m interested in stress related disorders. How are CRF receptors involved in the pathogenesis of stress-related disorders such as depression, metabolic syndrome, or in the long-term, neurodegenerative diseases such as Alzheimer’s disease? The levels of this receptor and its signaling function both in the brain and in the body is likely modulated between a healthy person and a severely stressed person. How does the level of this receptor change because that is crucial to the job that it’s doing in initiating these stress phenotypes? And the only reasonable way to look at the changes in levels of this receptor is to look at the changes in the level of both its transcript, the mRNA coding for this receptor, and its protein, in the context of its cellular environment. This spatial information, at subcellular-, cellular-, and organ-wide resolution, matters a great deal. If I can image the group of cells where the level of this receptor is changing within the context of the entire organ and also map where this group of cells projects throughout the entire organ, then I can hopefully piece together the biological relevance of this receptor at multiple scales – at the level of cellular plasticity, at the level of cell-cell signaling, and at the level of circuit function in the bigger picture of a disease.
So, if you knew all this, then you could develop new ways to treat —
— We first need to understand the process of developing addiction if you want to reverse that process. If the process for developing PTSD involves certain changes which may or may not include upregulation of this receptor, now you have a target to say, “This is a section of the brain where if we could downregulate its expression, maybe this could be a treatment for PTSD.” This is research that has been done over several decades but not much progress has been made, particularly from a pharmaceutical perspective. I am much more interested not just in pharmacological strategies, but in identifying maladaptive circuit behaviors, and then envisioning new technologies that could modify the signaling patterns that give rise to disease symptoms.
“We’ve created the science behind it, it works, and we can implement it, but how can we make it economically feasible?”
Jennifer Treweek
Give me some examples of the work you will be doing in your lab.
The chemistry for tissue engineering, better ways to image tissue and obtain a lot of molecular information on that tissue. Normally when you image tissue you’re getting one form of information – you’re getting the protein content, you’re looking at RNA, or single targets. Whereas I want more multidimensional information. I want the protein, the transcriptional profile, as well as the connectome – a comprehensive map of specific neural connections in the brain that reflect the particular disease state. So, I want to get lots of data out of the tissue I’m preparing and that requires some tissue engineering, to optimize the hydrogels, for example, to optimize the labeling techniques so you can get more information out of the tissue. Lots of classic systems neuroscience and also biomolecular engineering. Figuring out the targets for severe depression, or PTSD and other hard disease topics that are stress-related. We’re studying the pathogenesis of disease, the circuit changes that correlate with pathogenesis, and then looking at how we can reverse those changes in circuit activity. So if you see changes happening that are leading to the generation of a depressive state or addiction, how can we deliver modulation to reverse that? Right now, for Parkinson’s disease, for example, they’re doing this with deep brain stimulation, however they’re starting very late in the disease because they’re driving an electrode deep into someone’s brain so you don’t want to start early because you’re going through so many layers of tissue. But I want to help develop treatment modalities that involve manipulating circuits early on and hopefully much less invasively.

Jennifer Treweek hopes that one day the methods developed in her lab will become widespread and affordable so that everyone struggling from addiction, depression or PTSD can have access to better treatments with less averse effects. Photo/Daniel Druhora
We talked about a few areas of focus, but what is your big passion project?
To generalize about the U.S., people are very good at saying, “Okay, I have this short-term problem and I want a pill for it.” It’s a very short-sighted view of disease. I’m more interested in diseases that are much more complex, like depression, which looks very different in different patients. It’s so multifaceted in terms of the symptoms; you have metabolic symptoms, so now I want to study the effects of the CRF receptors in the gut and pancreas; it comes with sleep disorders and problems with memory and concentration. You have all of these symptoms in different tissues and organs in the body. My lab wants to look at these in a more body-wide approach to identify the circuits that are relevant. So I want to identify circuits of disease that go awry. From this understanding, we will be able to see how we can then modulate these circuits to slowly pull you out of the disease, symptom by symptom.
Let’s get out of the lab for a moment and imagine the future. What does the ideal future look like for you and your work?
We now have a roadmap for personalized medicine. We can take a patient’s cancer cells and come back with a more personalized way to treat it. But that is not economically feasible. We’ve created the science behind it, it works, and we can implement it, but how can we make it economically feasible? It should be deployed at a worldwide scale and not just for people who can come to Cedars Sinai or City of Hope for treatment. The same economic barriers will certainly come into play as we develop new technologies for better treating PTSD or Alzheimers’s disease, and these technologies probably won’t be in the form of a single pill, but instead ways of targeting circuits one-by-one so as to address symptoms one-by-one. It will be great to see how all of these technologies come together not just to create better treatments for disease, but also how we are able to translate these technologies into broadly accessible lean, real world treatments.
Published on September 17th, 2019
Last updated on May 14th, 2025