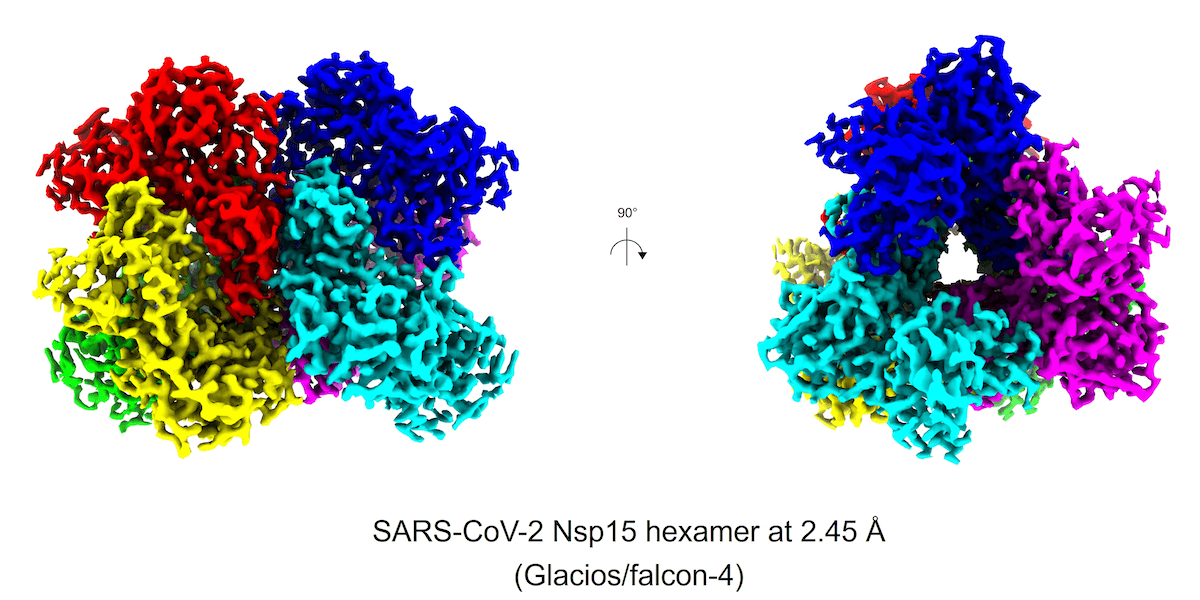
Cryo-EM image of the COVID virus
The image we’ve all now come to recognize as the COVID virus (SARS-CoV-2) – the sphere with the crown-like spikes going off in every direction – that image allowed structural biologists to understand the 3D structure of the virus at a near-atomic level, which led to the rapid development of the COVID-19 mRNA vaccines.
And that image was only possible thanks to cryogenic electron microscopy (cryo-EM).
Since 2022, USC has been home to a state-of-the-art cryo-EM facility at the USC Michelson Center for Convergent Bioscience in the Core Center of Excellence in Nano Imaging.
The USC cryo-EM facility comes from a partnership with biopharmaceutical research company Amgen and is part of an effort to increase the speed of scientific advances through partnerships between academic research facilities and for-profit corporations. The facility can be used by academic researchers from USC and other institutions, as well as corporate partners.
But in order for it to be used, it needed to be easily usable, and a few computational challenges had to be addressed. This is why researchers at USC Viterbi’s Information Sciences Institute (ISI) and the team at the USC Center for Advanced Research Computing (CARC) were brought in to provide automated scientific workflow solutions.
Scientific Workflow – What Is It and How Do You Automate It?
A scientific workflow is a way to describe a series of computational tasks and the dependencies among them. The defining property of a scientific workflow: it manages data flow. An example might be: “retrieve data from a microscope; reformat data; then run an analysis on reformatted data.”
Pegasus is an open source software developed at ISI that maps and automates complex workflows. It’s a National Science Foundation funded project led by ISI Principal Scientist Ewa Deelman. Pegasus is both an ongoing research project at ISI, since 2001, and a product that is used by a broad community of researchers spanning disciplines like astronomy, bioinformatics, earthquake science and gravitational-wave physics. As of 2022, structural biology with cryo-EM can be added to that list.
Mats Rynge and Karan Vahi, computer scientists from Deelman’s ISI team, were brought onto the cryo-EM team to automate the system using Pegasus.
Vahi said, “There are some partial software solutions available for cryo-EM processing, but none fit the integration requirements and the performance we needed.” Fortunately for USC, the ISI team was ready to tackle the computational challenges with Pegasus.
The Computational Challenges of Cryo-EM
Terabytes of Data
A quick primer on how cryo-EM works: a sample is flash-frozen; electrons are bounced off the sample to form an image; this is repeated at random orientations to create tens of thousands of 2D images; then the 2D images are combined to reveal the 3D structure of the sample.
All that bouncing and all those images create huge amounts of data – orders of magnitude greater than what is generated by a traditional microscope (think terabytes). And that data needs to be transferred to a facility where it can be processed in order for it to be useful.
This is where the USC Center for Advanced Research Computing (CARC) led by Byoung-Do Kim, a state-of-the-art computing facility on campus, comes into the picture. CARC operates high-performance computing (HPC) clusters that are well suited for processing these large datasets.
Rynge and Vahi collaborated with Kim and CARC staff member Tomasz Osinski to develop a comprehensive computational environment that could process terabytes of data from the cryo-EM facility using the CARC HPC resources.
End-to-End Computation
“The USC cryo-EM facility is one of only four cryo-EMs in the country that offers an end-to-end computation environment,” said Kim. This means that the data automatically goes from the cryo-EM facility to the CARC HPC for processing – two physically separated facilities.
How would that work without Pegasus? Kim said, “Researchers [at institutions without end-to-end computation] run their session with cryo-EM and the data is collected on the computer at the microscope. They have to copy the data to an external hard drive or transfer it directly to their own computer or simulation system, then launch the simulation. So, every step requires human effort and is done manually.”
With Pegasus, that process is automated. Once the cryo-EM session begins, an automated program moves the data to the CARC central storage. Pegasus kicks in and processes the data in real-time as it is generated.
Kim said, “Pegasus takes the data as it comes. It keeps feeding the data into the pipeline and manages the whole workflow. Data transfer, job submission, pre-processing, and all the movements in between – that’s what Pegasus is doing.”
Initial Stages and a Feedback Loop
Another challenge of cryo-EM comes in the initial stages of the workflow. A researcher’s entire session might take up to 24 hours of non-stop data collection. If for some reason the sample is corrupted or the parameters aren’t correct, you don’t want to find that out after 24 hours of data collection. Ideally, you’d want to know right away.
Rynge and Vahi identified this as one of the areas where Pegasus could help.
Along with Kim, Osinski and CARC staff member Cesar Sul, they developed a preprocessing option, giving cryo-EM users the ability to start the preprocessing immediately after initiating the microscope session. The user receives real-time feedback enabling them to adjust parameters during data acquisition.
This means there’s no more waiting 24 hours just to see that your session wasn’t usable. In fact, CARC staff members Jimi Chu and James Hong developed an unprecedented user notification integration with the application Slack. This level of data transfer automation and app integration is not typically available in cryo-EM workflows and has proven invaluable for monitoring purposes. With Pegasus, the user receives a Slack message on their phone and is able to look at the preprocessing images and decide if they want to intervene remotely.
Existing Infrastructure
“A lot of the project was about tightly integrating with the existing infrastructure. Before USC had this microscope, we had our USC CARC accounts. We had the HPC. We had the data storage facility. Researchers had USC user IDs to log into an existing system. All of that was already in place,” said Rynge. “We wrote the workflow from scratch but this really came down to an integration project.”
And the team came up with innovative ways to integrate the cryo-EM facility with the existing USC campus computing infrastructure, as well as making the entire cryo-EM process user-friendly to researchers who might be unfamiliar with computer science.
The Non-Computer Scientists
The typical researcher using the USC cryo-EM facility is likely someone studying structural biology – the scientific field that looks at how biological components are built. In fact, an early focus of the USC facility has been to use cryo-EM to help structural biologists identify changes in molecules when they are presented with other molecules, such as new drugs.
These structural biologists are experts in their field, not necessarily in computer science. Rynge said, “The reason we’re doing all this is usability, it’s for a non-computer scientist to be able to use cryo-EM.” He continued, “The typical user doesn’t even know that Pegasus is behind this. They just know something is processing their images… and here they are.”
What’s Next for Pegasus and Cryo-EM?
Researchers at USC and beyond are making use of the cryo-EM facility for all their nano-imaging needs. A recent USC project used the facility to study HIV, revealing structural information about the virus that will help in the development of antiretroviral therapeutics.
In 2022, the cryo-EM-CARC-Pegasus team published An Automated Cryo-EM Computational Environment on the HPC System using Pegasus WMS and presented it at a IEEE/ACM Workshop held alongside the annual Supercomputing Conference (SC 2022).
And they continue to look for new opportunities to improve the system, promising “there is more to come” with Pegasus and cryo-EM.
Osinski has plans to update the pipeline: “While we have a functional system, there is still a lot to be done. The messages sent to users can have potentially many more features, for example, predicting the number of object particles, which can help assess if the object dilution is optimal.”
And Rynge and Vahi continue to support the software, working with the team to ensure the workflow is running smoothly and stays up to date. Its newest version, offering more automation and better performance, is about to be deployed.
Deelman said, “Putting Pegasus in the experimental loop is really exciting. Now Pegasus is processing data as it is coming off the instrument in time to provide feedback to users so that they can adjust their experiment. This opens up the opportunity to automate not only cryo-EM-based experimentation analysis but also other time-critical experiments.”
Published on April 17th, 2023
Last updated on May 16th, 2024