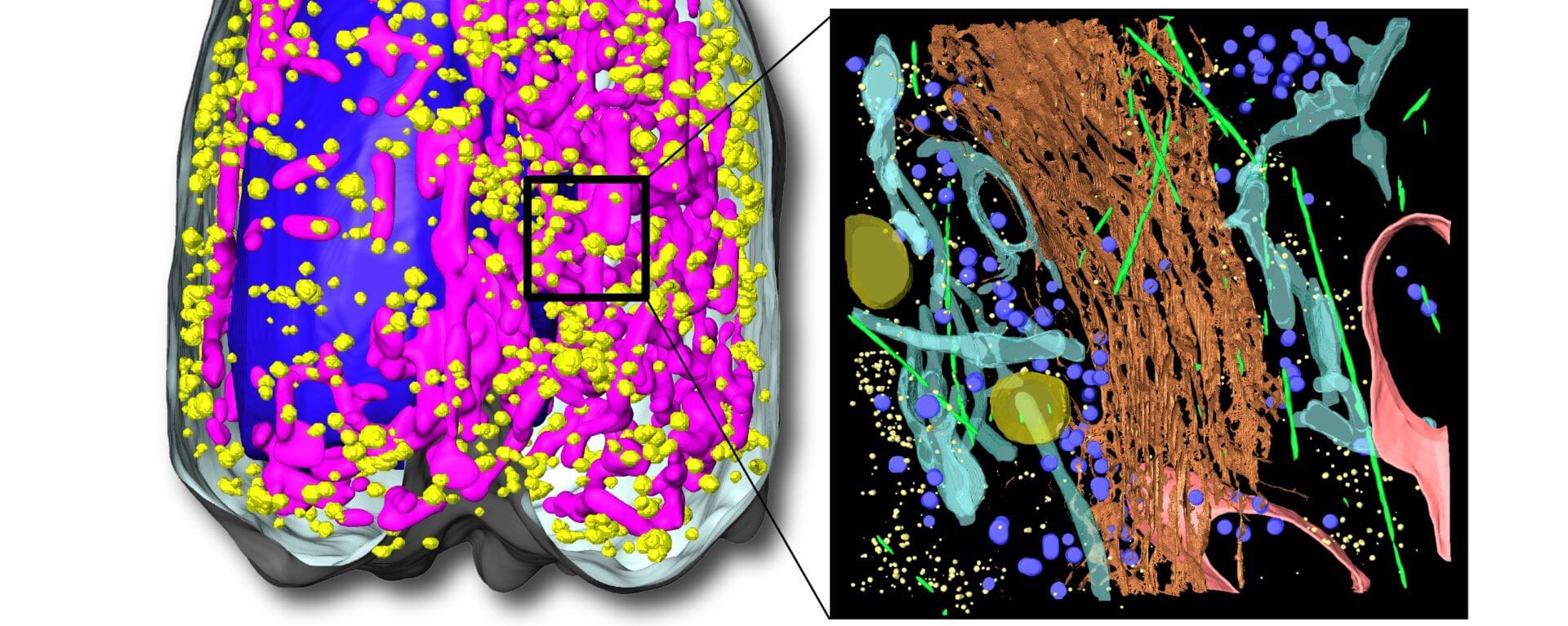
Mathematical modeling to can be used to understand the dynamics of cell function beyond what high resolution images can display. Photo credit: Kate White.
Cells are the basic building blocks of life. In every human body, trillions of them work together to perform the everyday functions necessary to survive. Despite how important and abundant they are, we know surprisingly little about how their inner functions work. This is for a very simple reason: they are too small to collect data in a reliable manner.
Using the limited information available, scientists usually create simulations of individual parts of the cell, or organelles. But these mathematics-based models were designed by different people for a vast variety of applications, so are often difficult to combine or compare. Researchers from the USC Michelson Center for Convergent Bioscience, in conjunction with colleagues at the University of California, San Francisco (UCSF) and ShanghaiTech University in China, have developed a system for combining these models into a compatible mathematical framework. They refer to it as a metamodel, or a model of models. The metamodel acts as a universal translator between the developed models for each organelle, providing information on how they communicate and how changes within one affect the other.
“The ultimate goal is to understand what’s in a cell, how all the different pieces talk to each other, and where they move over time,” explains Kate White, Gabilan Assistant Professor of Chemistry at the USC Dornsife College of Letters, Arts and Sciences. “If we are able to understand that, then we might be able to understand how things break down during disease.”
White is the director of the Pancreatic Beta Cell Consortium, a collection of scientists across the globe dedicated to the research of beta cells through a unique interdisciplinary approach. A number of these scientists’ work in metamodeling was recently published in the Proceedings of the National Academy of Sciences (PNAS), a prestigious academic journal.
“The goal of this consortium is not to model pancreatic beta cells just because they are interesting—even though they are very interesting,” says Barak Raveh, an independent principal investigator at Hebrew University of Jerusalem. “But if we can model these cells, we can model any cell. We can even model more complex systems, like all the organs in the body.”
Raveh is one of the chief author on the paper recently published in PNAS, titled “Bayesian metamodeling of complex biological systems across varying representations.”
He cited the interdisciplinary nature of the consortium as one of the main reasons for the project’s success.
“This collaborative environment is very unique in the world of science,” said Raveh. “We have people from very different scientific backgrounds: experimentalists, computational folks, artists, city designers. There is a certain Renaissance spirit to it all, where you must take into account perspectives completely different from your own.”
“This project is about building new modes of doing science.”
Carl Kesselman
The Pancreatic Beta Cell Consortium and its work with the USC Michelson Center have received widespread support and acclaim.
“We’re creating a community around cell modeling, where everyone depends on each other and the result that we obtain is fundamentally multi-disciplinary,” says Carl Kesselman, a professor of industrial and systems engineering, computer science, preventive medicine, and biomedical sciences at the USC Viterbi School of Engineering.
“I think of it as a three-legged stool, where the three legs are computational modelling, experimental techniques, and data management,” says Kesselman, the William M. Keck Chair in Engineering. “If you take away any of those pieces, the stool’s going to fall down.”
Thanks to the success of this initial project, the consortium plans to continue their cross-cultural, multidisciplinary approach to research. Now that the metamodel has been proven to work — Raveh calls this paper a “proof of concept” — the team will apply the metamodeling concept to delve into researching the various methods of communication between organelles within the cell. A broader understanding of this process can provide crucial breakthroughs in understanding how cells respond to disease outbreaks and medications.
The most immediate application of their research into pancreatic beta cells revolves around these cells’ capacity to produce insulin, which can revolutionize the treatment of diabetes. Theoretically, however, the method can be applied to a wide range of illnesses, as all of them will agitate the organelles in a cell in some manner. The framework the team has built could have ripple effects that spread over the entire field of medicine.
“Ultimately, this project is about building new modes of doing science with regards to how we manage, create, and share data, and how to make that transparent and reproducible,” says Kesselman. “The question we ask ourselves is: How can we build systems that transform how we think about doing science?”
Published on October 18th, 2021
Last updated on November 15th, 2022